CF520C.DNA Alignment
普及/提高-
通过率:0%
AC君温馨提醒
该题目为【codeforces】题库的题目,您提交的代码将被提交至codeforces进行远程评测,并由ACGO抓取测评结果后进行展示。由于远程测评的测评机由其他平台提供,我们无法保证该服务的稳定性,若提交后无反应,请等待一段时间后再进行重试。
题目描述
Vasya became interested in bioinformatics. He's going to write an article about similar cyclic DNA sequences, so he invented a new method for determining the similarity of cyclic sequences.
Let's assume that strings s and t have the same length n , then the function h(s,t) is defined as the number of positions in which the respective symbols of s and t are the same. Function h(s,t) can be used to define the function of Vasya distance ρ(s,t) :
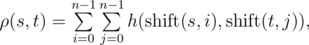 where
where  is obtained from string s , by applying left circular shift i times. For example, ρ("AGC","CGT")= h("AGC","CGT")+h("AGC","GTC")+h("AGC","TCG")+ h("GCA","CGT")+h("GCA","GTC")+h("GCA","TCG")+ h("CAG","CGT")+h("CAG","GTC")+h("CAG","TCG")= 1+1+0+0+1+1+1+0+1=6 Vasya found a string s of length n on the Internet. Now he wants to count how many strings t there are such that the Vasya distance from the string s attains maximum possible value. Formally speaking, t must satisfy the equation:
is obtained from string s , by applying left circular shift i times. For example, ρ("AGC","CGT")= h("AGC","CGT")+h("AGC","GTC")+h("AGC","TCG")+ h("GCA","CGT")+h("GCA","GTC")+h("GCA","TCG")+ h("CAG","CGT")+h("CAG","GTC")+h("CAG","TCG")= 1+1+0+0+1+1+1+0+1=6 Vasya found a string s of length n on the Internet. Now he wants to count how many strings t there are such that the Vasya distance from the string s attains maximum possible value. Formally speaking, t must satisfy the equation: 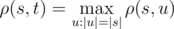 .
.
Vasya could not try all possible strings to find an answer, so he needs your help. As the answer may be very large, count the number of such strings modulo 109+7 .
输入格式
The first line of the input contains a single integer n ( 1<=n<=105 ).
The second line of the input contains a single string of length n , consisting of characters "ACGT".
输出格式
Print a single number — the answer modulo 109+7 .
输入输出样例
输入#1
1 C
输出#1
1
输入#2
2 AG
输出#2
4
输入#3
3 TTT
输出#3
1
说明/提示
Please note that if for two distinct strings t1 and t2 values ρ(s,t1) и ρ(s,t2) are maximum among all possible t , then both strings must be taken into account in the answer even if one of them can be obtained by a circular shift of another one.
In the first sample, there is ρ("C","C")=1 , for the remaining strings t of length 1 the value of ρ(s,t) is 0.
In the second sample, ρ("AG","AG")=ρ("AG","GA")=ρ("AG","AA")=ρ("AG","GG")=4 .
In the third sample, ρ("TTT","TTT")=27